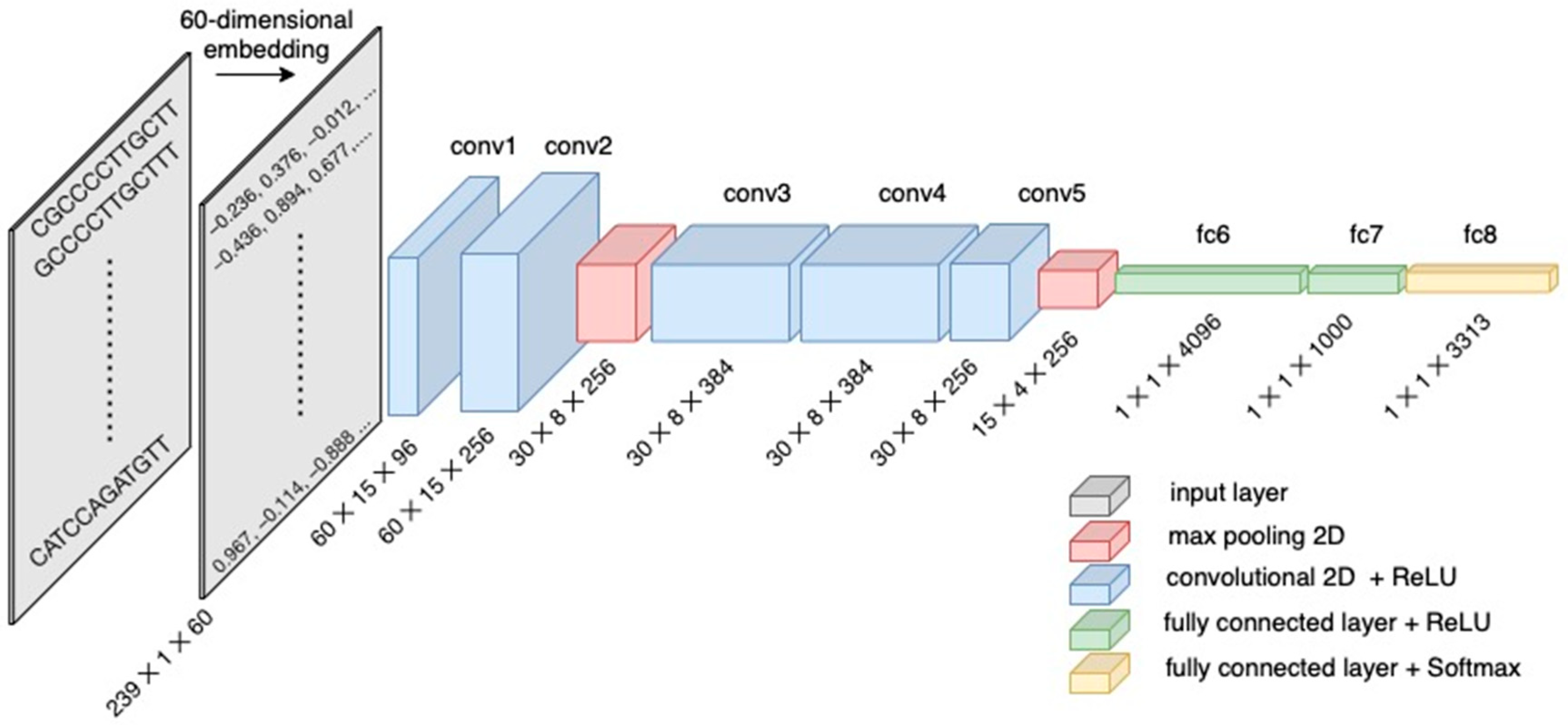
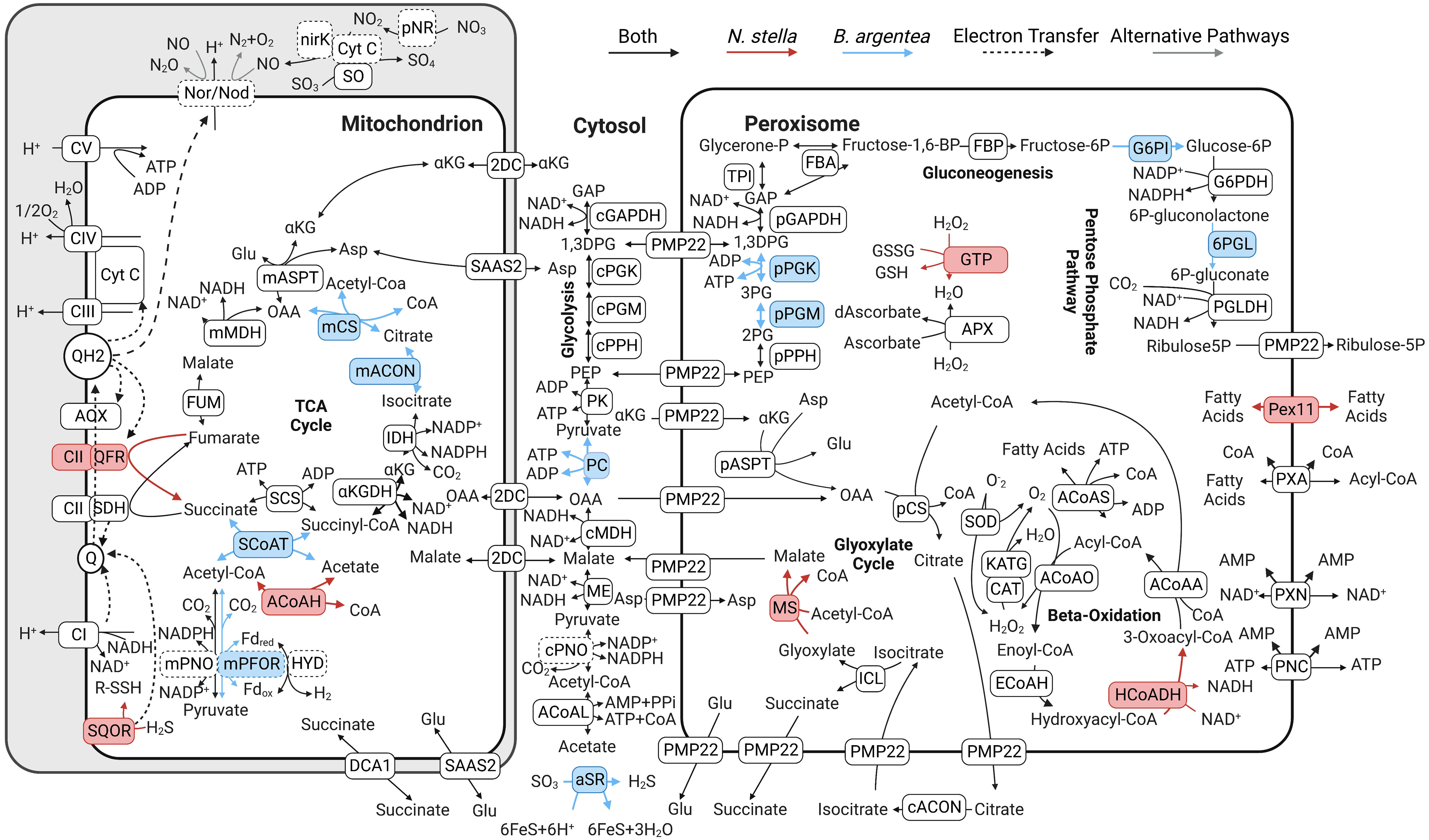
Welcome to Dr. Ying Zhang’s Laboratory of Computational Molecular Ecology
I am a computational biologist with broad interests in metabolic modeling, (Meta)omics, and other data intensive applications in environmental microbiology. My research group develop open source software and models for the simulation of biological systems, with a central objective of achieving mechanistic understandings at molecular, organismal, and ecosystems scales. Our recent applications include genome-scale modeling of non-model bacteria and archaea, (meta)transcriptomics and genomics of marine protists, metagenomics of free-living and host-associated microbiomes, and taxonomic classification of sequencing reads using deep learning neural networks.